Performance Optimization When Using rxExec() to Parallelize Algorithms
Microsoft R Server is designed around the concept of PEMAs (Parallel External Memory Algorithms). An external memory algorithm is an algorithm which does not require all data to be present in memory, it can process data sequentially, generating intermediate results and then iterating on those results. These algorithms are widely used for statistical analysis and machine learning, and allow large datasets to be computed on relatively small amounts of memory. PEMAs are implementations of these algorithms which allow chunks of data to be processed in parallel on many cores of the system. Such algorithms require the separation of the requisitely sequential code segments from the parallelizable segments.
The rxExec() function in Microsoft R Server is not, in and of itself a PEMA, but is a tool provided to the user for the writing of parallel algorithms. Out of the box parallel MRS Algorithms, such as rxBtrees() or rxLinMod(), to name a few, are intrinsically designed to run on various distributed systems. rxExec() allows users to parallelize algorithms for operation on many cores of a single system; and allows algorithms which were not designed to operate on distributed data process data on distributed systems.
Another important thing to note is Microsoft R Server operates as a closed source package within the open source R interpreter. This is achieved by using an open source, Microsoft maintained, library: RxLink. RxLink mitigates passing data between the open source R process(es) and the closed source Microsoft R Server process(es) via named pipes. (for details, please see: Importing and Exporting Large In-Memory Dataframes). This mode of operation leads us to make performance versus ease-of-use considerations when writing custom PEMAs using rxExec(); requiring us to pay attention to how we manage marshalling data from open source R to Microsoft R Server. rxExec() takes as its [required] arguments:
- FUN, this function will be executed on nodes of a distributed system or cores of a local machine. The configuration of this execution depends on the active compute context. Setting compute context will be briefly demonstrated in this article.
- “…” or elemArgs, if arguments are specified as a comma separated list, they will be passed to the parallelized function and used on every execution of the function. If ElemArgs is specified, it must be a list whose length matches the number of parallelizations of an algorithm, the list will be iterated as every parallel operation is started, and pass each unique input to a separate instance of the function. Both depend on the arguments required by said function, this can be empty. elemArgs usage will be demonstrated in this article.
- elemType, this defines the distributed computing mode to use, and the allowable types depend on the compute context. The default is "nodes" for a distributed compute context, which tells rxExec() to execute as one parallel execution per distributed node, and "cores" for a local compute context, which tells rxExec() to execute as one parallel execution per logical core.
A common use of rxExec() is to define a custom function for execution and passing the function, as well as its input parameters, to rxExec(), which will be provided to each parallel execution of the function.
For the purpose of generality, and providing examples which everyone can try, this article will focus on the local parallel compute context, “localpar”, that is, using rxExec() to parallelized algorithms across many cores of your local machine.
Let’s examine a simple example. We will use some open source packages to define a random dataset, and use rxExec() to take sums of subsets of the data in parallel, using system.time() to measure execution speed. In the first part of the example, we will use rxExec() the "naïve" way. Afterwards we will make some optimizations to our script, and utilize rxExec() in a more "correct" way. We will compare the performance and explain optimizations that can help the user maximize their performance.
Example:
# Open source package for creating data tables, will be used to create random data
library(data.table)
# Open source package That we will use to verify the results of rxExec()
library(foreach)
# Used by foreach to iterate through an elements of a vector list
library(iterators)
# Our dataset will have 5 million rows
nrow <- 5e6
# Create a data table with 5 million row, 3 columns (A random letter between ‘A’
# and ‘G and two random deviates based on the row number)
test.data <- data.table(tag = sample(LETTERS[1:7], nrow, replace = T), a = runif(nrow), b = runif(nrow))
# Run the following line if you wish to verify the dimensions of the created table
# are 5 million by 3
dim(test.data)
# Next, we want to get all unique values in the first column, there is not guarantee
# that all letters A – G were used
tags <- unique(test.data$tag)
# Now lets define a function to take the subset of rows, based on the value in the first
# column, and take the sum of data column “a” of all of these rows
subsetSums <- function(data, tags) {
x <- subset(data, tag == tags)
sum(x$a)
}
# First let’s run this using Foreach, which will iterate through every tag sequentially
# and take the sums of the provided data columns.
resForeach <- foreach( tag = iter(tags) ) %do% {
subsetSums (test.data, tag)
}
# We’re going to use rxExec() to parallelize the above function, running each sum on a core of our machine
# rxExec is wrapped in system.time() to measure it’s performance, cat('MKL Threads=',getMKLthreads(),'\n')
# will tell us how man simultaneous sums we can perform. Breaking down the rxExec() call, we have specified
# the function “subsetSums”, and it’s two input parameters, data getting test.data, and tags getting tags
# wrapped in rxElemArg() which will give each parallel execution of “subsetSums” its own tag to operate on
system.time({
cat('MKL Threads=',getMKLthreads(),'\n')
rxSetComputeContext("localpar")
resRxExec <- rxExec(subsetSums, test.data, rxElemArg(tags))
})
When we execute this code, we see the following execution times (in seconds):
| User: | 14.06 |
| System: | 26.56 |
| Elapsed: | 74.00 |
#To ensure our data is consistent between the two, we test with:
identical (resRxExec, resForeach)
and the result is:
[1] TRUE
This was the naïve way to use rxExec(): Write a function and then call that function, passing the input parameters, within rxExec().
Unfortunately, we saw less than stellar performance. Considering the volume of data, this is not terrible, but, we can do much better with a few simple optimizations.
The fundamental error with our non-optimized code is that we ignore how Microsoft R Server works inside of open source R. Because Microsoft R Server is a separate process from R itself, they do not have a shared memory space, all messages between the main R process and the Microsoft R Server process must be sent via named pipes.
To send a message across a named pipe, the RxLink library will serialize data contained within the R process's memory and send it to Microsoft R Server, after Microsoft R Server has complete operating on data in its memory space, it will serialize the results and send them back to RxLink for deserialization.
if we examine our call to rxExec(), we can see the bottleneck which is degrading our performance:
# subsetSum takes as input data and tags. Tags are relatively small, but data is a copy of the entire dataset.
subsetSums <- function(data, tags) {
x <- subset(data, tag == tags)
sum(x$a)
}
# Keeping in mind that test.data is the entire dataset, we can see that buy passing test.data, for every parallel call of subsetSums
# a copy of the entire dataset must be serialized by RxLink and sent from the R process via named pipe to the Microsoft R Server
# process in which rxExec() is executing. This serialization deserialization is a huge overhead, and the reason we get subpar performance
# from our parallelization
system.time({
cat('MKL Threads=',getMKLthreads(),'\n')
rxSetComputeContext("localpar")
resRxExec <- rxExec(subsetSums, test.data, rxElemArg(tags))
})
To optimize this parallel algorithm, we want to remove the overhead caused by serialization/deserialization. To do this, we will change how susbsetSums() accesses the dataset.
Example, the setup segment of the code is identical:
# Open source package for creating data tables, will be used to create random data
library(data.table)
# Open source package That we will use to verify the results of rxExec()
library(foreach)
# Used by foreach to iterate through an elements of a vector list
library(iterators)
# Our dataset will have 5 million rows
nrow <- 5e6
# Create a data table with 5 million row, 3 columns (A random letter between ‘A’ and ‘G’ and two random deviates based on the row number)
test.data <- data.table(tag = sample(LETTERS[1:7], nrow, replace = T), a = runif(nrow), b = runif(nrow))
# Next, we want to get all unique values in the first column, there is not guarantee that all letters A – G were used
tags <- unique(test.data$tag)
# Now, we will save the dataset to an Rdata object, which will be used to circumvent the serialization bottleneck.
# We will measure the execution time of this, factoring it into our overall performance time. For more information
# on saveRDS(), please run “?saveRDS” in R (Before running the demo <path\\to\\folder> must be changed to a valid
# path)
system.time({
saveRDS(test.data, "<path\\to\\folder>\\data.Rdata", compress = FALSE)
})
# Now lets define a new version of subsetSums, this version does not take data as an input, but, instead, reads it
# directly. For more information on readRDS, please execute “?readRDS” in R. (Before running the demo <path\\to\\folder>
# must be changed to a valid path)
subsetSums <- function(tags) {
data <- readRDS("<path\\to\\folder>\\data.Rdata")
x <- subset(data, tag == tags)
sum(x$a)
}
# First let’s run this using Foreach, which will iterate through every tag sequentially and take the sums of the provided data columns.
# Note that we no longer specify test.data in the call to subsetSums()
resForeach <- foreach( tag = iter(tags) ) %do% {
subsetSums (tag)
}
# Just as last time, we’re going to call rxExec(), except this time, test.data is not an input to subsetSums(), this prevents RxLink from serializing the
# entire dataset and sending copies to every instance of subsetSums() executing within Microsoft R Server, instead, each instance reads the data
# directly, which, as we will see, has a significant impact on performance
system.time({
cat('MKL Threads=',getMKLthreads(),'\n')
rxSetComputeContext("localpar")
resRxExec <- rxExec(subsetSums, rxElemArg(tags))
})
Executing this script, we get the execution time of as (in seconds):
| saveRDS() | + | rxExec() | = | Total | |
| User: | 0.70 | 0.02 | 0.72 | ||
| System: | 0.14 | 0.05 | 0.19 | ||
| Elapsed: | 0.86 | 9.72 | 10.58 |
Which is performance improvement of:
| User: | 98.47% | (19.53x Faster) |
| System: | 99.28% | (139.79x Faster) |
| Elapsed: | 88.10% | (6.99x Faster) |
Viewed graphically, this is:
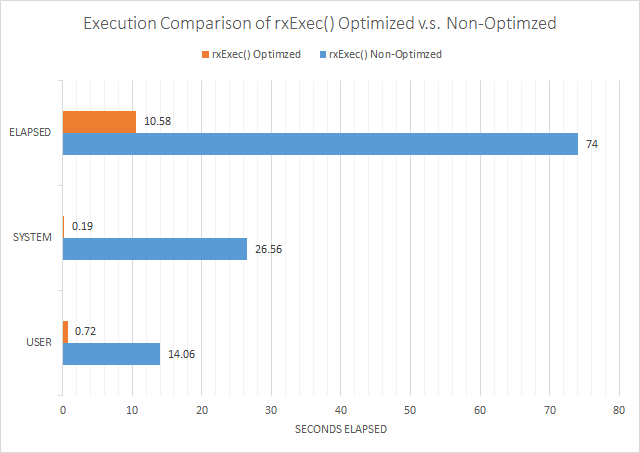
In conclusion, the performance bottleneck of transmitting large chunks across the named pipe is staggering, and completely defeats the speed increase that you get from rxExec()'s ability to parallelize algorithms. When writing R scripts for Microsoft R Server, a user cannot ignore the overhead of data marshalling between open source R and Microsoft R Server.